SOX8
Research Question
This task-based tutorial builds upon three previous tool-oriented tutorials build_bedpe, query_bedpe, and plot_contacts and also introduces an alternate solution using get_loops. Becoming familiar with these tools is recommended before diving into this tutorial. In this tutorial, we will explore two different approaches to answer the following questions:
For the SOX8 gene, which TSS-CpG pairings show the strongest and weakest acetylation signal in 3D space?
To answer this question, we will determine acetylation levels by assessing and visualizing the changes in H3K27ac HiChIP reads between DMSO (control) and dCBP1 (treatment) in the RH4 cell type. We will use bedtools to obtain the SOX8 TSS, the TAD in which SOX8 resides, as well as all the CpG sites within the TAD. Next, we will use build_bedpe to construct pairings between the SOX8 TSS and all CpG sites within the TAD, and determine contact frequencies of the pairings using query_bedpe. We will use plot_contacts to visualize the process and conclude with visualizations of both the strongest and weakest acetylation signals in our case vs control analysis.
To list all available data on the Tinker box, simply type the following command into the terminal window:
list_samples.shLet’s assign simple names to our control, treatment, and reference files:
control=RH4_DMSO
treatment=RH4_HDACi
TSS=~/lab-data/hg38/reference/gencode.TSS.200bp.hg38.bed
TAD=~/lab-data/hg38/reference/TAD_goldstandard_lift.hg38.bed
CPG=~/lab-data/hg38/reference/cpg.hg38.bed
Obtain TSS
Now that we have a TSS file ready, we can obtain the SOX8 TSS
cat $TSS | grep SOX8 > SOX8_tss.bedObtain TAD
Using the SOX8 TSS, we can fetch the TAD it resides in
bedtools intersect \
-a SOX8_tss.bed
-b $TAD \
-wa -wb
| cut -f 6-8 > SOX8_tad.bed
cat SOX8_tad.bed
chr16 790000 1259999Obtain CpG sites
We now have the SOX8 TSS and the TAD it resides in. Using the SOX8 TAD, let’s now obtain all the CpG sites housed in it.
bedtools intersect \
-a $CPG
-b SOX8_tad.bed > SOX8_cpgs.bedVisualize SOX8 TAD
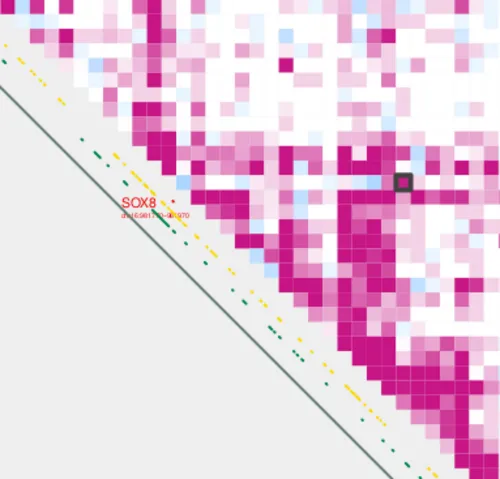
We can now visualize the SOX8 TAD using plot_contacts. CpG sites and TSSs will be displayed by default.
plot_contacts.sh \
--sample1 $control \
--range chr16:790000:1260000:SOX8_TAD\
--genome hg38 \
--output_name SOX8.pdf \
--norm aqua
--max_cap 2 \
--resolution 5000
--annotations_default TRUE \
--annotations_custom SOX8_tad.bed \
--annotations_custom_color 657c78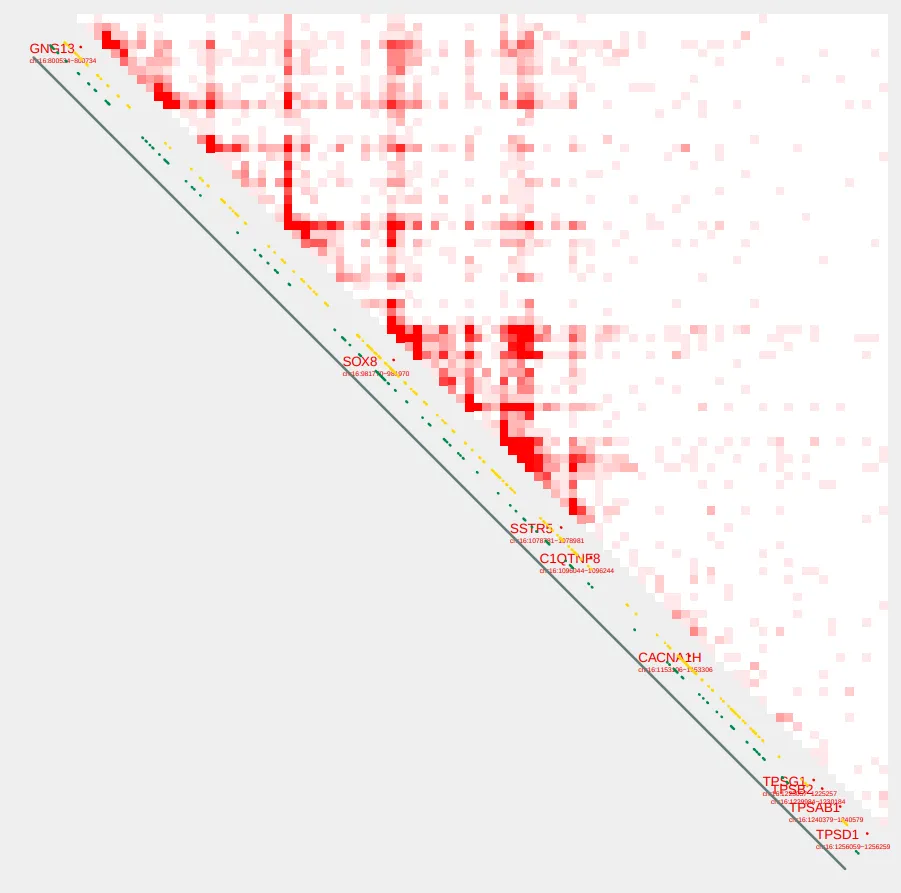
Let’s zoom in to SOX8.
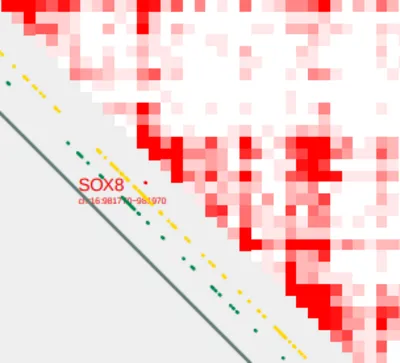
Construct Pairs
Next, we can construct pairings between the SOX8 TSS and all CpG sites within the TAD by using build_bedpe.
build_bedpe.sh \
-A SOX8_tss.bed \
-B SOX8_cpgs.bed > SOX8_tss_cpg.bedpe
bedpe=SOX8_tss_cpg.bedpeVisualize pairs
We can visualize these pairings using the --bedpe option with plot_contacts
plot_contacts.sh \
-sample1 $control \
-range chr16:790000:1260000:SOX8_TAD \
-genome hg38 \
-output_name SOX8_bedpe.pdf \
-norm aqua \
-resolution 5000 \
-max_cap 2 \
-bedpe $bedpe \
-bedpe_color 424242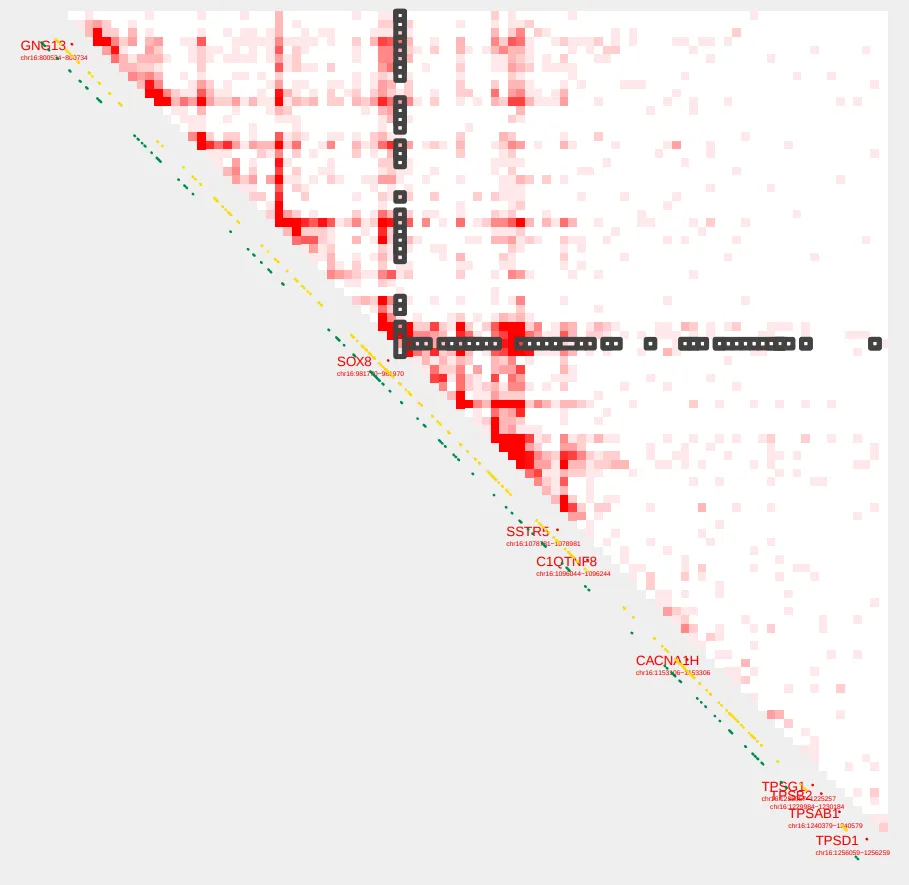
Obtain and sort contact frequencies
Great! We now have all the genomic pairings we want! We can now obtain the contact frequencies of these pairings using query_bedpe and sort them in increasing order
query_bedpe.sh \
--sample1 $control \
--sample2 $treatment \
--pair SOX8_tss_cpg.bedpe \
--genome hg38 --norm aqua --resolution 5000 \
| sort -k9n > SOX8_tss_cpg_contact_values.bedpeWe can view the top and bottom values using head and tail commands. Since we sorted our list in increasing order, head will show us the smallest acetylation values and tail will show us the largest acetylation values.
head -n 5 SOX8_tss_cpg_contact_values.bedpe
chr16 979878 985327 chr16 981770 981970 0.745 0 -0.745
chr16 807341 808025 chr16 981770 981970 1.117 0.379 -0.738
chr16 817032 817833 chr16 981770 981970 0.559 0 -0.559
chr16 869858 870117 chr16 981770 981970 0.559 0 -0.559
chr16 800227 800806 chr16 981770 981970 0.372 0 -0.372
tail -n 5 SOX8_tss_cpg_contact_values.bedpe
chr16 981770 981970 chr16 1089732 1089937 0 0.758 0.758
chr16 981770 981970 chr16 986820 987448 2.607 3.411 0.804
chr16 970219 971303 chr16 981770 981970 0.931 1.895 0.964
chr16 981770 981970 chr16 1050626 1050848 1.489 2.653 1.164
chr16 981770 981970 chr16 1057607 1057865 0 1.895 1.895Visualize case-control pairs
Before we visualize the lowest and highest acetylation pairs, let’s visualize all of our pairs again, but this time we’ll look at both control and treatment samples using the two-sample version of plot_contacts
plot_contacts.sh \
--sample1 $control \
--sample2 $treatment \
--range chr16:790000:1260000:SOX8_TAD \
--resolution 5000 \
--genome hg38 \
--output_name SOX8_delta_all.pdf \
--norm aqua \
--max_cap 2 \
--annotations_default TRUE \
--annotations_custom SOX8_tad.bed \
--annotations_custom_color 657c78 \
--bedpe SOX8_tss_cpg.bedpe \
--bedpe_color 424242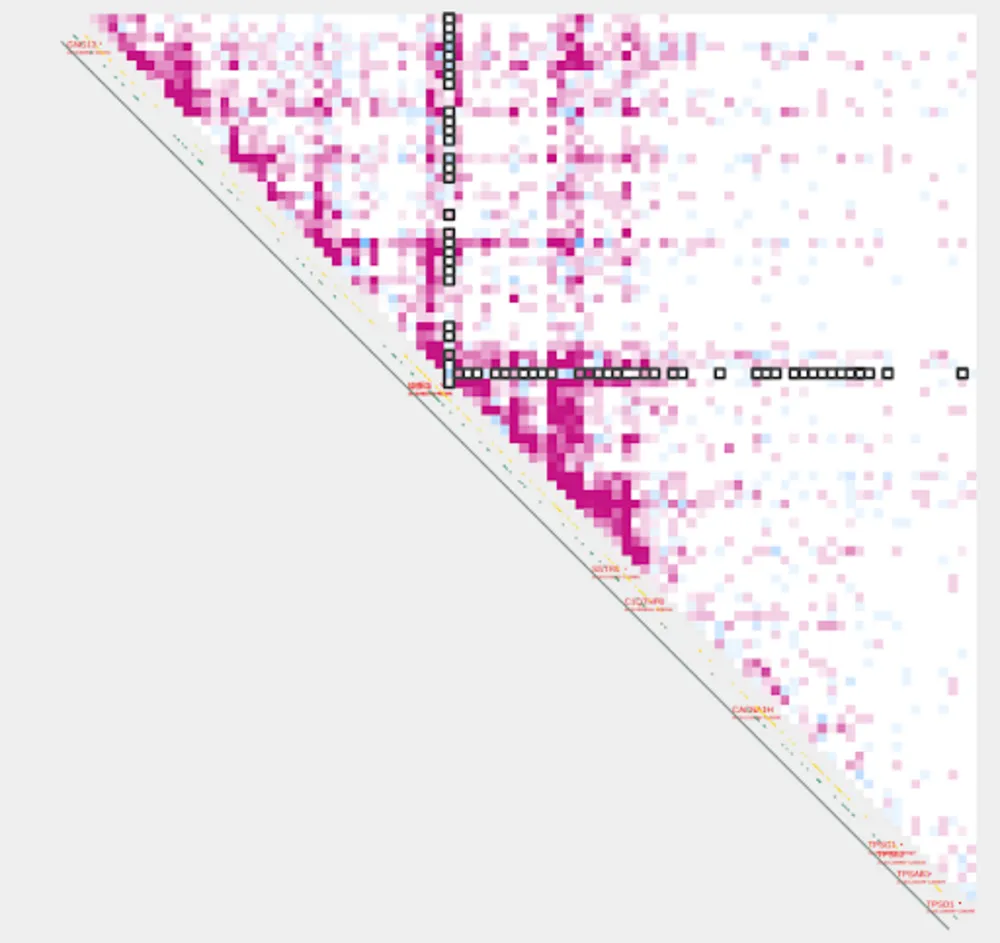
Save pairs with lowest and highest acetylation
Let’s save the pair with the lowest acetylation:
head -n 1 \
SOX8_tss_cpg_contact_values.bedpe > low_SOX8_tss_cpg_contact_values.bedpeAnd let’s also save the pair with the highest acetylation:
tail -n 1 \
SOX8_tss_cpg_contact_values.bedpe > high_SOX8_tss_cpg_contact_values.bedpeVisualize lowest and highest acetylation pairs
Now we can plug these bedpe files into the --bedpe option of plot_contacts to visualize the pairs with the lowest and highest acetylation values in our case vs control analysis.
plot_contacts.sh \
--sample1 $control \
--sample2 $treatment \
--range chr16:790000:1260000:SOX8_TAD \
--resolution 5000 \
--genome hg38 \
--output_name SOX8_delta_low.pdf \
--norm aqua \
--max_cap 2 \
--annotations_default TRUE \
--annotations_custom SOX8_tad.bed \
--annotations_custom_color 657c78 \
--bedpe low_SOX8_tss_cpg_contact_values.bedpe \
--bedpe_color 424242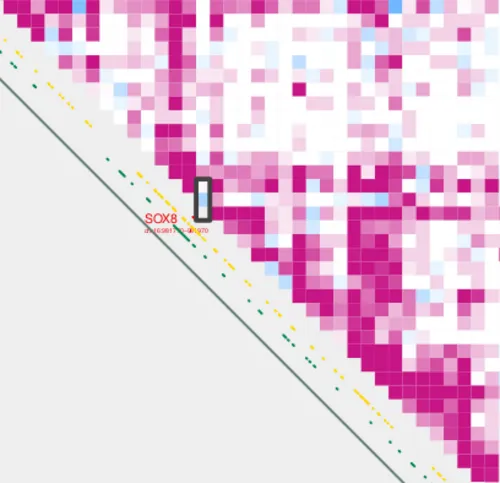
plot_contacts.sh \
--sample1 $control \
--sample2 $treatment \
--range chr16:790000:1260000:SOX8_TAD \
--resolution 5000 \
--genome hg38 \
--output_name SOX8_delta_high.pdf \
--norm aqua \
--max_cap 2 \
--annotations_default TRUE \
--annotations_custom SOX8_tad.bed \
--annotations_custom_color 657c78 \
--bedpe high_SOX8_tss_cpg_contact_values.bedpe \
--bedpe_color 424242